| Original Article | ||
Open Vet. J.. 2022; 12(4): 551-561 Open Veterinary Journal, (2022), Vol. 12(4): 551–561 Original Research A survey of contagious ecthyma and molecular characterization of Orf virus in sheep and goats in Nigeria (2014–2016)Adeyinka Jeremy Adedeji1*, Jolly Amoche Adole1, Olayinka Oluwafemi Asala1, Ahmed Abdulkadir Gamawa2, Nanven Abraham Maurice1 Anvou Jambol1, Mohammed Bashir Bolajoko1, Nneka Chineze Chima1, Victoria Isioma Ifende1, Yiltawe Simwal Wungak1, Timothy Yusufu Woma1,3, and Pam Dachung Luka11National Veterinary Research Institute, Vom, Nigeria 2Animal Health Technology Department, Bauchi State College of Agriculture, Bauchi, Nigeria 3The Pirbright Institute, Woking, United Kingdom *Corresponding Author: Adeyinka Jeremy Adedeji. National Veterinary Research Institute, Vom, Nigeria. Email: yinkadeji [at] yahoo.com Submitted: 29/09/2021 Accepted: 14/07/2022 Published: 17/08/2022 © 2022 Open Veterinary Journal
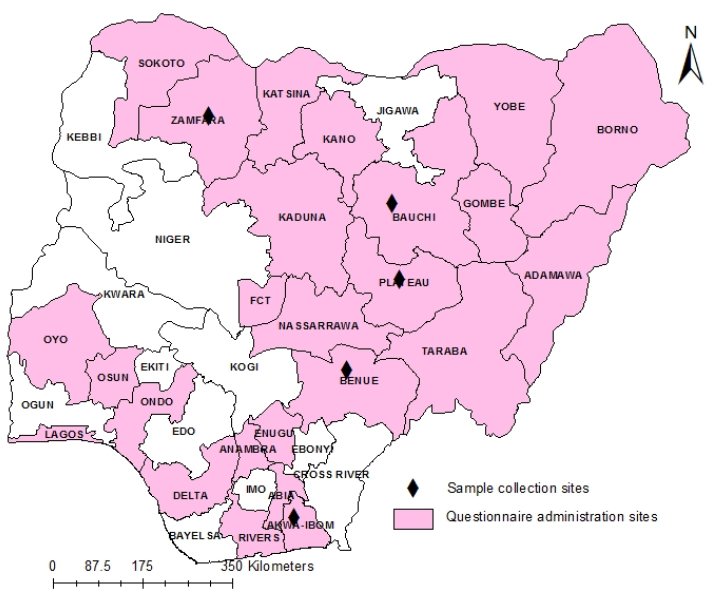
AbstractBackground: Outbreaks of contagious ecthyma (CE) are frequently reported in sheep and goat flocks in Nigeria with severe clinical outcomes. CE is a debilitating and economically important disease primarily affecting sheep and goats caused by the Orf virus (ORFV). Despite field reports of CE in the country, there is no concise country-wide epidemiological data on the disease and limited genetic data of circulating Nigerian ORFV are available in the public domain. Aim: An epidemiological survey of CE and molecular characterization of ORFV circulating in Nigeria from 2014 to 2016. Method: Data were collected using designed questionnaires, administered to veterinarians and farmers in selected States of Nigeria. Samples were collected during passive surveillance for CE from 2014 to 2016 which were analyzed by polymerase chain reaction (PCR). The A32L and B2L genes of circulating ORFV were also characterized. Results: Analysis of the questionnaire showed that 69.54% (n =82/118) of the farmers claimed to have experienced CE in their flocks with average morbidity and mortality rates of 25% and 15%, respectively. A total of 113 veterinarians participated in the study, with 69.9% (n =79) familiar with CE and claimed CE causes morbidity rates of 25%–37% and mortality rates of 10%–15% in sheep and goats. Laboratory results revealed that ORFV was detected in 72% (18/25) of outbreak samples analyzed by real-time PCR. Phylogenetic analysis of A32L and B2L genes revealed that Nigerian ORFV sequences belong to clusters I and II and are similar to viruses from India, Ethiopia, and China. Conclusions: This study is the first nationwide epidemiological data on the status of CE in sheep and goats in Nigeria. It is also the first report of molecular characterization of two genes of ORFV circulating and causing outbreaks in small ruminants in the country. This study showed that CE is under-reported, widespread and of economic importance to sheep and goat farmers in Nigeria. Keywords: A32L gene, B2L gene, Contagious ecthyma, Orf virus, Small ruminants. IntroductionContagious ecthyma (CE) or Orf is a debilitating and economically important disease of sheep and goats caused by the epitheliotropic Orf virus (ORFV) a member of the genus Parapoxvirus and family Poxviridae (Spyrou and Valiakos, 2015). The ORFV primarily affects sheep and goats but has been reported in cattle, camels, and wild animals (Nandi et al., 2011; Adedeji et al., 2018a; Şevik, 2019). The genome of ORFV is linear double-stranded DNA with an estimated size of 134–139kb and it is the smallest member within the subfamily Chordopoxvirinae (Friederichs et al., 2014). Genetically, the A32L and B2L genes encode highly conserved segments of the ORFV, which have been used for detection and phylogenetic studies of the virus (Chan et al., 2009). The A32L gene of ORFV encodes an ATPase that mediates virion DNA packaging, while the envelope gene (B2L) is the highly immunogenic protein of the virus (Gelaye et al., 2016). CE is a zoonotic disease and occupational hazard to farmers, butchers, and veterinarians handling livestock (Essbauer et al., 2010; Veraldi et al., 2019; Andreani et al., 2019). Clinically, the disease is characterized by fever, papules, pustules, and thick tenacious scabs at oral commissures, lips, mouth, nose, ears, legs, genital region, and udders (Nandi et al., 2011). These skin lesions become ulcerative and painful, with the affected animals unable to either feed or walk, leading to weight loss and reduced productivity (Bala et al., 2018). However, CE is also characterized by high morbidity and low mortality, but, in young animals, mortality is high due to the inability to suckle (Windsor et al., 2017). ORFV is transmitted by contact through damaged skin during grazing and abrasions developed on the lips, nostrils, and mouth caused by dried feeds or plants (Spyrou and Valiakos, 2015). The ORFV is distributed worldwide, particularly in sheep and goat-producing countries (Nandi et al., 2011; Bala et al., 2018). Diagnosis of CE is based on clinical signs and laboratory techniques such as electron microscopy, viral isolation, and molecular assays (Torfason and Guðnadóttir, 2002; Kottaridi et al., 2006; Chan et al., 2009). Since the first report of CE in Nigeria in 1978, reported outbreaks of the disease have resulted in economic losses to sheep and goat farmers in the country (Odo, 2003; Adedeji et al., 2018b). Despite, reports of CE outbreaks in some parts of Nigeria with varying economic losses (Adedeji et al., 2017; Ifende et al., 2019). There is a paucity of epidemiological data on the disease nationally and limited genetic sequences of the circulating ORFV in Nigeria are available in the public domain. Albeit, only one sequence from one gene of ORFV circulating in Nigerian small ruminants flock has been deposited in the public domain or reported (Lawal et al., 2021). Therefore, an epidemiological survey was carried out to determine the status of CE in Nigeria and characterized the circulating ORFV causing outbreaks. Materials and MethodsStudy areaNigeria is located in West Africa and divided into 36 states. There are five major agro-ecological zones in the country consisting of humid (rainforest), sub-humid (southern Guinea savannah, Northern Guinea Savannah), semi-arid (Sudan Savannah), and arid (Sahel Savannah) zones (Fadiga et al., 2013). There are two main climatic seasons in Nigeria, namely; Rainy season (May to September) and dry season (October to April). The rainy season extends to October-December in the southern coastal states of the country (Fadiga et al., 2013). Nigeria has a population of 42 million sheep and 73 million goats (Bolajoko et al., 2019). Keeping sheep and goats is an integral part of the lives of rural households in Nigeria. These animals are kept for food, source of income, and religious purposes and serve as “bank” for saving money (Bolajoko et al., 2019). Most of the sheep and goats in Nigeria are reared under an extensive or semi-intensive system of management. The common breeds of sheep are Yankasa, Udah, and Balami reared mostly in sub-humid, semi-arid, and arid agro-ecological zones. The West African dwarf (WAD) sheep are reared mostly in humid zones (Ngere et al., 1984; Fadiga et al., 2013). In contrast, the common breeds of goats in Nigeria are Sahel, Sokoto Red and Kano Brown found in the sub-humid and semi-arid and arid agro-ecological zones. While WAD goats are mostly kept in the sub-humid and humid zones (Fadiga et al., 2013; Yusuf et al., 2018). Field data collectionIn this study, epidemiological data on CE was collected by administering standardized and pretested questionnaires to veterinarians, and farmers using interview, conducted in the selected study areas (Fig. 1). For veterinarians, the questionnaires were administered at their Annual National Conference in November 2016. Out of 130 questionnaires administered, 113 veterinarians from 25 of the 36 states of Nigeria returned their filled questionnaires (Fig. 1). While 118 farmers from 7 states in Nigeria participated in the study (Fig. 1), the interviews were conducted between February and May 2017. Purposive sampling methodology was used, and the availability of the veterinarians and farmers to be interviewed was utilized. The questionnaire for veterinarians focused on diagnosis/identification of CE, age, sex, breeds of sheep and goats affected by CE, and season of the year with the highest incidence of CE. Other information obtained were the cost of management, morbidity rates, mortality rates, and economic importance of CE. Information from livestock farmers was collected using structured interviews with the aid of pictures showing lesions of CE for easier understanding. The questions were translated into local languages as appropriate. The interview focused on the livestock management system, familiarity with CE, age, and occurrence of CE outbreak in their flock in the last year before the interview. Other questions include factors associated with CE outbreaks, morbidity/mortality rates and breeds of the sheep and goats mostly affected by CE. Questions were also asked on the economic importance of disease such as the effect of CE on the trade value of affected animals. Sample collectionScab samples were collected from suspected CE outbreaks in different parts of Nigeria (Fig. 1). Twenty-five samples were submitted from 2014 to 2016, and samples from the same flocks were pooled together. The samples were collected from a livestock market, abattoirs, and backyard smallholder flocks from five states in Nigeria (Fig. 1). The samples were submitted to the Virology Division, National Veterinary Research Institute, Vom, Nigeria for laboratory analysis. Laboratory analysisThe scab samples were homogenized, and genomic DNA extracted using QIAamp® DNA Mini kit (QIAGEN, Hilden Germany) following the manufacturer's instructions. ORFV freeze-dried vaccine (Onderstepoort, South Africa) was used as positive control and subjected to DNA extraction. All the samples were analyzed using a polymerase chain reaction (PCR) assay for the detection of eight poxviruses of livestock based on high-resolution melting curve analysis (HRMCA) (Gelaye et al., 2017). The panel of controls were ORFV, Pseudocowpox virus (PCPV), GTPV, SPPV, Lumpy skin disease virus (LSDV), Bovine papular stomatitis virus, Cowpox virus (CPXV), and Camel pox virus. The positive control panels for the PCR assay were provided by the Animal Production and Health Section, Joint Food and Agriculture Organization-International Atomic Energy Agency, Seibersdorf, Vienna, Austria. Positive samples were characterized by amplification and sequencing of A32L and B2L genes fragments of the ORFV using primers as previously described (Gelaye et al., 2016).
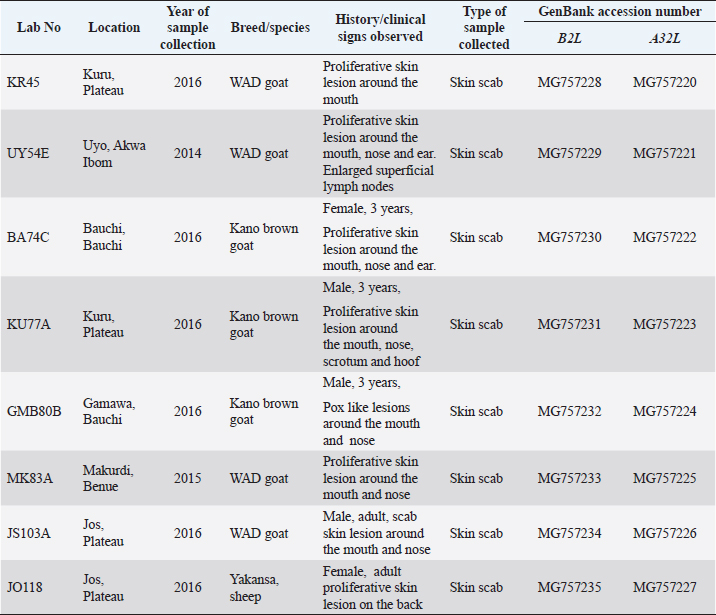
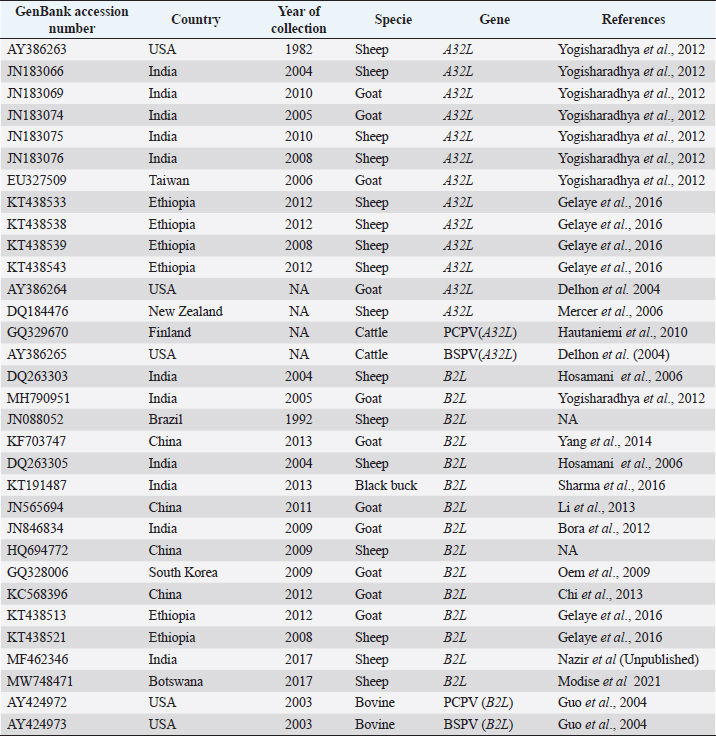
Fig. 1. Map of Nigeria showing states where the questionnaire survey was carried out and samples collected. Data analysisData obtained from the questionnaires were entered into a spreadsheet and analyzed using descriptive statistics and tabulated. Phylogenetic analysis of A32L and B2L genes of ORFVEight ORFV positive samples were selected showing spatial temporal spread for molecular characterization targeting two genes of the virus (Table 1). The A32L gene of ORFV encoding an ATPase that mediates virion DNA packaging and the B2L envelope gene which are the highly immunogenic proteins were charactered using the Sanger sequencing method (Chan et al., 2009; Gelaye et al., 2016). ORFV sequences were generated commercially by Macrogen Inc. (South Korea). The raw sequence data were assembled using Staden Package (http://staden.sourceforge.net/) and multiple alignments were performed using the MUSCLE condon algorithm implemented in MEGA X (Kumar et al., 2018). Sequences of the two gene segments of ORFV were compared to available sequences in the GenBank using the Basic Local Alignment Search Tool (BLAST) (https://blast.ncbi.nlm.nih.gov/Blast) at default settings. MEGA X was used for phylogenic analysis by Neighbor-joining tree inferred using p-distance method and data resampled 1,000 times in bootstrap method (Kumar et al., 2018).To construct the phylogenic trees, 13 A32L and 15 B2Lgene sequences representing the two known ORFV clusters as well as the sequences of BSPV and PCPV were retrieved from GenBank, and listed in Table 2 (Gelaye et al., 2016). The nucleotide sequences generated in this study were deposited in the NCBI GenBank sequence database with accession numbers MG757220–MG757227 for the A32L gene and MG757228–MG757235 for the B2L gene (Table 1). Table 1. List of Nigerian ORFV from field outbreaks and their GenBank accession numbers.
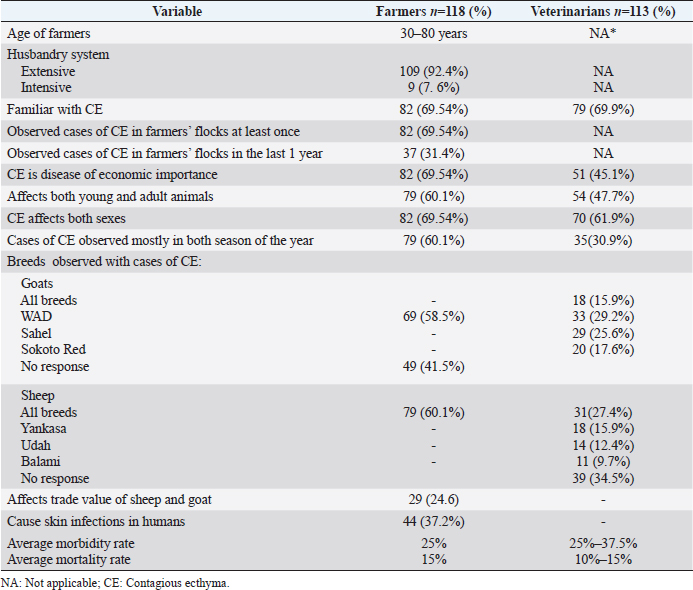
Ethical approvalThis study was approved by National Veterinary Research Institute Animal Ethics Committee (AEC/03/58/18). ResultsQuestionnaire data analysisSheep and goats farmers interviewed were from seven states of Nigeria (Akwa Ibom, Bauchi, Kano, Kaduna, Plateau, Yobe, Zamfara) (Fig. 1). The farmers interviewed were of ages 30–80 years old and 92.4% (n =109/118) of them practice extensive husbandry system of livestock of farming (Table 3). Data from the survey also revealed that 69.54% (n =82/118) of the farmers were familiar with CE and had observed CE outbreaks in their flocks at least once. Additionally, 31.4% (n=37/118) of the farmers said they had observed cases of CE in their flocks in the last year before the interview was conducted. Also, 69.54% (n =82/118) of the farmers indicated that CE is a disease of economic importance in Nigeria. Similarly, the analyzed data showed that 60.1% (n =79/118) of the farmers indicated that CE affects both young and adult animals, while 69.54% (n =82/118) of farmers claimed CE affects sheep and goats of both sexes. Furthermore, 60.1% of the farmers claimed that CE occurs during both dry and rainy seasons. Regarding breed of goats mostly affected, 58.5% (n=69/118, Table 3) stated that WAD as the breed of goats mostly affected by CE. At the same time, all the respondents did not identify any breed of sheep as most susceptible to CE. Furthermore, 24.6% (n =29) of farmers claimed CE affects the trade price of animals with the disease. Farmers also indicated that the average morbidity and mortality rates of CE were 25% and 15% respectively during outbreaks in sheep and goats. Additionally, 37.2% (n =44) of farmers claimed that CE causes skin infections in humans. A total of 113 veterinarians participated in the questionnaire survey from 25 states of Nigeria (Table 3). Amongst the veterinarians that participated in the study, 69.9% (n =79) were familiar with CE. In comparison, 45.1% (n =51) said CE is of economic importance with average morbidity of 25% in sheep and 37.5% in goats, respectively, as well as average mortality of 10% in sheep and 15% in goats. Concerning breed of goats, 29.2% (n =33) of veterinarians claimed the WAD as the most affected goats while others indicated Red Sokoto (25.6%, n =29) and Sahel (17.6%, n =20) as other breeds affected by CE in Nigeria. The data collected from the questionnaire survey revealed that 47.7% (n =54) of veterinarians indicated that CE affects sheep and goats of all ages, while 61.9% (n =70) said CE affects both male and female animals. Regarding the season, 30.9 % (n =35) of veterinarians stated that CE occurs mainly during the rainy season. The veterinarians’ questionnaire survey also revealed that the average cost of managing a CE outbreak is estimated to be $2 per animal (sheep or goat). Table 2. List of B2L and A32L gene sequences of ORFV and other parapoxviruses used for phylogenetic analysis.
Table 3. Summary of epidemiological data collected from questionnaire survey on CE of sheep and goats in Nigeria.
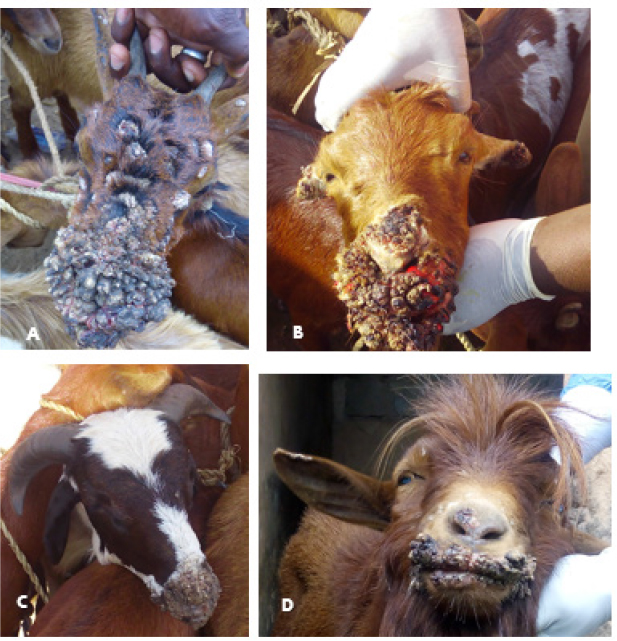
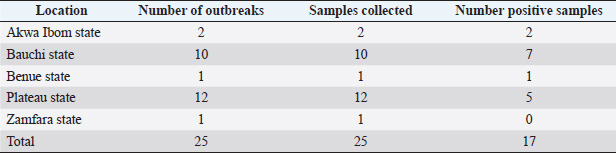
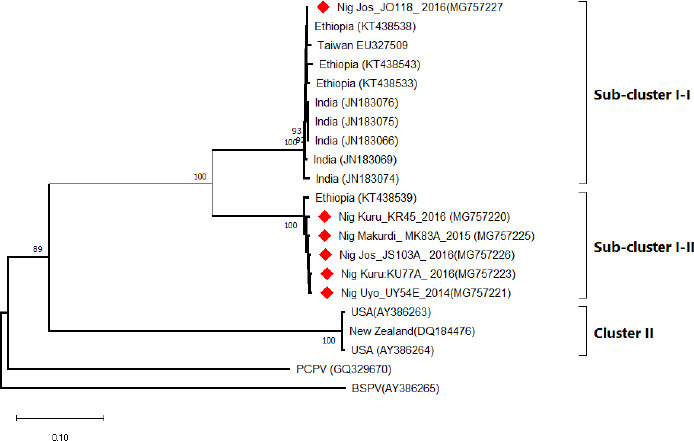
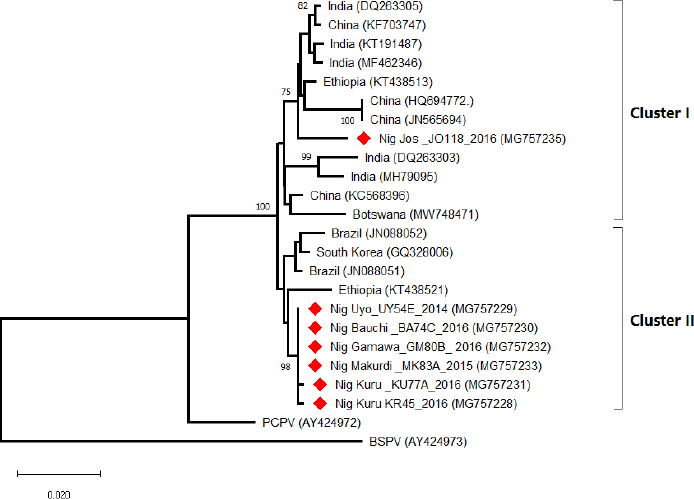
Field observations and laboratory resultsDuring field investigation of CE outbreaks, clinical signs observed were proliferative scab lesions around the mouth, noses, mammary glands, and hoofs (Table 1, Fig. 2A–D). Twenty-five samples were collected from sheep and goats in the following states of Nigeria, namely, Akwa Ibom, Benue, Bauchi, Plateau and Zamfara (Fig. 1, Table 4). The samples were collected from Kano Brown (Fig. 1 A, B, and D) and WAD breeds of goats and Yankasa sheep, (Fig. 2C). ORFV was detected by multiplex Real-time PCR in 72% (18/25) of the samples analyzed. SPPV, GTPV, LSDV, PCPV, and CPXV were not detected by the multiplex Real-time PCR in the samples analyzed. Phylogenetic analysis of A32L and B2L genes of ORFVPhylogenetic analysis of A32L gene segment of Nigerian ORFV showed 7 Nigerian ORFV A32L gene sequences (KR45, UY54E, BA74A, KU77A, MK83 and JS103) with 96%–98% similarity and clustering in Cluster I, sub-cluster I and II (Fig. 3). Meanwhile, ORFV sample JO118 grouped Cluster I together with ORFVs from Ethiopia, India and Taiwan. A32L gene Sub-cluster II of Cluster I was the dormant and widespread ORFV in Nigeria between 2014 and 2016, detected in Akwa Ibom, Bauchi, Benue, and Plateau states (Fig. 3). The B2L gene segment of seven of Nigerian ORFV isolates (KR45, UY54E, BA74A, KU77A, MK83, and JS103) had 97%–99% similarity. The Nigerian ORFV isolates grouped with Cluster II when inferred on the phylogenic tree with ORFVs from Brazil, Ethiopia, and South Korea (Fig. 4). While, ORFV isolate JO118 grouped with Cluster I together with ORFVs from Botswana, China Ethiopia, and India. DiscussionThis study is the first nationwide epidemiological data collection on the status of CE in sheep and goats in Nigeria. It is also the first report of molecular characterization of two genes of ORFV circulating and causing outbreaks in small ruminants in the country. Previously, only case reports and studies at the regional level were documented for CE in Nigeria without a countrywide concise study of the disease (Adedeji et al., 2017; Ifende et al., 2019). In this study, most veterinarians (69.9%), farmers (69.4%), and butchers (64.03%) that participated in the questionnaire survey were familiar with CE, which suggests that cases of the disease frequently occur in various parts of the country. It also implies that CE outbreaks may be widespread in Nigeria. Interestingly, based on findings in this study, a higher percentage of farmers considered CE a disease of economic importance compared to veterinarians (Table 3). Also, farmers claimed average mortality rates of 15% as a result of CE outbreaks in flocks of sheep and goats which is high, particularly for poor rural communities in Nigeria that depend on livestock for their livelihood. Recent studies in parts of Nigeria and Laos reported mortality rates of 33%–100% in smallholder goats flocks (Windsor et al., 2017; Adedeji, et al., 2018b). This further suggests that CE may be causing greater economic losses, but is underreported by farmers and neglected by relevant authorities. Besides, CE incidence rates of 10%–90% have also been reported in some parts of the world (Bala et al., 2018). Concerning the economic importance of CE, majority of farmers and veterinarians interviewed stated that CE is of economic importance. A study conducted in Nigeria also reported that traders and farmers agreed that CE affects the trade prices of affected sheep and goats (Gambo et al., 2018). In rural communities in Nigeria, it is common practice for livestock farmers to quickly sell their sick animals directly to livestock traders or butchers to mitigate losses during disease outbreaks (Bolajoko et al., 2019). In another study in Nigeria, 7,258 cases of CE in sheep and goats were reported during ante mortem inspections over 9 years at an abattoir (Ifende et al., 2019). This finding suggests that farmers sell animals showing clinical signs of CE to butchers for slaughter without reporting to relevant veterinary authorities. Most veterinarians in this study claimed that CE affects goats more than sheep, which agrees with a report in South Africa with a similar finding (Scagliarini et al., 2012). In addition, veterinarians and farmers claimed a higher incidence of CE occurs in WAD goats than other breeds of goats in Nigeria. Likewise, our earlier study in Nigeria suggested a similar finding (Adedeji et al., 2018b). The laboratory result confirms that CE outbreaks occur in Nigeria based on analysis with the multiplex real-time PCR. Due to the possibility of co-infections and the similarity of CE with other poxvirus infections of sheep and goats, a multiplex real-time PCR was used to analyse the samples. All the samples were negative for sheep pox and goat pox-viruses. The ORFV was detected in samples collected from four out of five states. Before this study, CE had not been reported in Bauchi and Benue states of Nigeria. The phylogenetic analysis of the A32L and B2L gene showed the diversity of the ORFV circulating in Nigeria, which clustered with sequences of ORFV from Brazil, Ethiopia, China, and India (Figs. 3 and 4). All the Nigerian ORFV isolates in this study cluster together apart from sample JO118_Nig, which was collected at a livestock market in Jos Plateau State. ORFV belonging to cluster I, sub-cluster II, and cluster II based on the two gene sequences analysed were the most dominant strain of the virus circulating in Nigeria. This is the first report of the phylogenetic analysis of two gene fragments of ORFV circulating in Nigeria. Although CE is not a notifiable disease, however, based on the data collected in this study, farmers have expressed concern about the impact of the disease on their flocks.
Fig. 2. (A): Proliferative lesions of CE around the mouth, nose and eyes of a Kano brown goat at a livestock market in Bauchi, Bauchi state. (B): Proliferative lesions of CE around the mouth, nose and eyes of a Kano brown goat in Bauchi, Bauchi state. (C): Scab lesions of CE on the mouth and nose of Yankasa ram in Gamawa, Bauchi state. (D): A Kano brown with proliferative lesions of CE in Kuru, Plateau. Table 4. Distribution of samples collected during CE outbreaks and laboratory results in five states of Nigeria from 2014 to 2016.
Fig. 3. The Phylogenic tree of A32L gene of the ORFV samples collected from outbreaks in Nigeria. The Nigerian ORFV samples are marked with red. The tree was constructed using the Neighbor-joining method at 500 bootstrap.
Fig. 4. The Phylogenic tree of the B2L gene of the ORFV collected from outbreaks in Nigeria. The Nigerian ORFV samples are marked with red. The tree was constructed using the neighbor-joining method at 1,000 bootstrap. ConclusionsThis study provides the first concise country-wide epidemiological data on CE in sheep and goats in Nigeria. It is also the first molecular characterization of two gene fragments of ORFV circulating and causing outbreaks in small ruminants. Findings from this study suggest that CE is an economically important disease of sheep and goats in Nigeria. It is, therefore, recommended that further studies be undertaken to investigate the impact of the disease on sheep and goat farming in the country. Conflict of interestThe authors declare that there is no conflict of interest. FundingThe authors of this study did not receive any form of funding. Authors’ contributionsAJA, MBB TYW, and VII designed the study, AJA, JAA, AAG, and NAM collected the data and samples. AJA, MBB, AJ, JAA, NCC, YSW, and PDL analysis data and samples. AJA, OOA, PDL, TYW wrote the manuscript. All authors read and approved the manuscript. AcknowledgmentsThe authors hereby acknowledge veterinarians, farmers, and abattoirs workers that participated in this study. The authors acknowledge Adrian Maguda, Dyek Yohonna Dyek, Dr. Ronke Odita, and Dr. David Lazarus who donated the positive controls. The authors also acknowledge the staff of Animal Production and Health Laboratory, Joint FAO/IAEA Laboratories Seibersdorf, Vienna Austria for HRMCA reagents used for the study. ReferencesAdedeji, A.J., Gamawa, A.A., Chima, N.C., Ahmed, A.I., Ifende, V.I., Adole, J.A., Ahmad, I., Woma, T.Y. and Luka, P.D. 2018. First report of camel contagious ecthyma in Nigeria. Open Vet. J. 8(2), 208–211. Adedeji, A.J., Adole, J.A., Chima, N.C., Maguda, A.S., Dyek, Y.D., Jambol, A., Anefu, E., Shallmizhil, J.J. and Luka, P.D. 2018. Contagious ecthyma in three flocks of goats in Jos-South, Plateau State, Nigeria. S. J. Vet. Sc. 16(1), 107–112. Adedeji, A.J., Maurice, N.A., Wungak, Y.S., Adole, J.A., Chima, N.C., Woma, T.Y., Chukwuedo, A.A. and Shamaki, D. 2017. Diagnosis of orf in West African Dwarf goats in Uyo, Akwa Ibom state, Nigeria. Afri. J. Infec. Dis. 11(2), 90–94. Andreani, J., Fongue, J., Bou Khalil, J.Y., David, L., Mougari, S., Le Bideau, M., Abrahão, J., Berbis, P. and La Scola, B. 2019. Human Infection with Orf Virus and description of its whole genome, France, 2017. Emerg. Infect. Dis. 25(12), 2197–2204. Bala, J.A., Balakrishnan, K.N., Abdullah, A.A., Mohamed, R.B., Haron, A.W., Jesse, F.F. and Mohd-Azmi, M.L. 2018. The re-emerging of orf virus infection: a call for surveillance, vaccination and effective control measures. Microb. Pathog. 120, 55–63. Bolajoko, M.B., Adedeji, A.J., Dashe, G.D., Òsemeke, O.H. and Luka, D. 2019. Molecular epidemiology and economic impact of goat pox on small holder sheep and goats farmers in North Central Nigeria. Small Rumin. Res. 179, 75–78. Bora, D.P., Barman, N.N., Das, S.K., Bhanuprakash, V., Yogisharadhya, R., Venkatesan, G., Kumar, A., Rajbongshi, G., Khatoon, E., Chakraborty, A. and Bujarbaruah, K.M. 2012. Identification and phylogenetic analysis of orf viruses isolated from outbreaks in goats of Assam, a northeastern state of India. Virus Gen. 45(1), 98–104. Chan, K.W., Yang, C.H., Lin, J.W., Wang, H.C., Lin, F.Y., Kuo, S.T., Wong, M.L. and Hsu, W.L. 2009. Phylogenetic analysis of parapoxviruses and the C-terminal heterogeneity of viral ATPase proteins. Gene 432(1–2), 44–53. Chi, X., Zeng, X., Hao, W., Li, M., Li, W., Huang, X., Wang, S. and Luo, S. 2013. Heterogeneity among orf virus isolates from goats in Fujian Province, Southern China. PLoS One 8(10), e66958. Delhon, G., Tulman, E.R., Afonso, C.L., Lu, Z., de la Concha Bermejillo, A., Lehmkuhl, H.D., Picone, M.E., Kutish, G.F. and Rock, D.L. 2004. Genomes of the parapoxviruses orf virus and bovine popular stomatitis virus. J. Virol. 78(1), 168–177. Essbauer, S., Pfeffer, M. and Meyer, H. 2010. Review Zoonotic poxviruses. Vet. Microbiol. 140(1), 229–236. Fadiga, M., Jost, C. and Ihedioha, J. 2013. Financial costs of disease burden, morbidity and mortality from priority livestock diseases in Nigeria: disease burden and cost–benefit analysis of targeted interventions. ILRI Res. Rep. Nairobi, Kenya pp: 1–84. Fleming, S.B., Wise, L.M. and Mercer, A.A. 2015. Molecular genetic analysis of orf virus: a poxvirus that has adapted to skin. Viruses 7(3), 1505–1539. Friederichs, S., Krebs, S., Blum, H., Wolf, E., Lang, H., von Buttlar, H. and Büttner, M. 2014. Comparative and retrospective molecular analysis of parapoxvirus (PPV) isolates. Virus Res. 181, 11–21. Gambo, P., Maguda, A.S., Adole, J.A., Dyek, Y.D., Ifende, V.I., Bot, C. and Adedeji, A.J. 2018. A survey of viral diseases of livestock characterized by skin lesions in Kanam Local Government Area of Plateau State, Nigeria. Nig. Vet. J. 39(3), 250–262. Gelaye, E., Achenbach, J.E., Jenberie, S., Ayelet, G., Belay, A., Yami, M., Loitsch, A., Grabherr, R., Diallo, A. and Lamien, C.E. 2016. Molecular characterization of orf virus from sheep and goats in Ethiopia, 2008–2013. Virol. J. 13(34); doi: 10.1186/s12985-016-0489-3. Gelaye, E., Mach, L., Kolodziejek, J., Grabherr, R., Loitsch, A., Achenbach, J. E., Nowotny, N., Diallo, A., & Lamien, C. E. 2017. A novel HRM assay for the simultaneous detection and differentiation of eight poxviruses of medical and veterinary importance. Sci. Rep. 7, 42892; doi: 10.1038/srep42892. Guo, J., Rasmussen, J., Wünschmann, A. and de la Concha-Bermejillo, A. 2004. Genetic characterization of orf viruses isolated from various ruminant species of a zoo. Vet. Microbiol. 99, 81–92. Hautaniemi, M., Ueda, N., Tuimala, J., Mercer, A.A., Lahdenpera, J. and McInnes, C.J. 2010. The genome of pseudocowpoxvirus: comparison of a reindeer isolate and a reference strain. J. Gen. Virol. 91, 1560–1576. Hosamani, M., Bhanuprakash, V., Scagliarini, A. and Singh, R.K. 2006. Comparative sequence analysis of major envelope protein gene (B2L) of Indian oruses isolated from sheep and goats. Vet. Microbiol. 116(4), 317–324. Ifende, V.I., Maurice, N.A., Abbas, Y., Agu, C., Bolajoko, M.B., Jambol, A., Adole, J.A., Asala, O., Wungak, Y.S., Maguda, A., Umeh, E. and Adedeji, A.J. 2019. A retrospective study of viral skin diseases of cattle, sheep and goats in Plateau State, Nigeria. S. J. Vet. Sci. 17(1), 49–55. Kottaridi, C., Nomikou, K., Lelli, R., Markoulatos, P. and Mangana, O. 2006. Laboratory diagnosis of contagious ecthyma: comparison of different PCR protocols with virus isolation in cell culture. J. Virol. Methods 134(1–2), 119–124. Kumar, S., Stecher, G., Li, M., Knyaz, C. and Tamura, K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. Lawal, N., Ibrahim, M., Onawala, D. A., Bello, M. B., Aliyu, R. M., Baraya, Y. S., Aliyu, A., Ibrahim, A. M., & Sa'adu, A. 2021. Molecular characterization and phylogenetic analysis of orf virus isolated from goats in Sokoto metropolis, Nigeria. Fut. Sci. OA. 7(6), FSO700; doi: 10.2144/fsoa-2020-0162. Li, H., Zhu, X., Zheng, Y., Wang, S., Liu, Z., Dou, Y., Li, H., Cai, X. and Luo, X. 2013. Phylogenetic analysis of two Chinese orf virus isolates based on sequences of B2L and VIR genes. Arch. Virol. 158(7), 1477–1485. Modise, B.M., Settypalli, T.B.K., Kgotlele, T., Xue, D., Ntesang, K., Kumile, K., Naletoski, I., Nyange, J.F., Thanda, C., Macheng, K.N., Marobela-Raborokgwe, C., Viljoen, G.J., Cattoli, G. and Lamien, C.E. 2021. First molecular characterization of poxviruses in cattle, sheep, and goats in Botswana. Virol. J. 18(1), 167. Mercer, A.A., Ueda, N., Friederichs, S.M., Hofmann, K., Fraser, K.M., Bateman, T. and Fleming, S.B. 2006. Comparative analysis of genome sequences of three isolates of orf virus reveals unexpected sequence variation. Virus Res. 116(1–2), 146–158. Nandi, S., Ujjwal, K.D. and Chowdhury, S. 2011. Current status of contagious ecthyma or orf disease in goat and sheep—A global perspective. Small Rumin. Res. 96(1–2), 73–82. Ngere, I.O., Adu, I.F. and Okubanjo, I.O., 1984. The Indigenous Goats of Nigeria. FAO/UNDP. In Animal genetic resources information. Rome, Italy: FAO (Food and Agricultural Organization of the United Nations), vol. 3, pp: 1–9. Odo, B.I. 2003. Comparative study of some prevalent diseases of ecotype goats reared in southeastern Nigeria. Small Rumin. Res. 50, 203–207. Oem, J.K., Roh, I.S., Lee, K.H., Lee, K.K., Kim, H.R., Jean, Y.H. and Lee, O.S. 2009. Phylogenetic analysis and characterization of Korean orf virus from dairy goats: case report. Virol. J. 6, 167. Scagliarini, A., Piovesana, S., Turrini, F., Savini, F., Sithole, F. and McCrindle, C.M. 2012. Orf in South Africa: endemic but neglected. Onderstepoort J. Vet. Res. 79(1), 1–8. Şevik, M. 2019. Orf virus circulation in cattle in Turkey. Comp. Immunol. Microbiol. Infect. Dis. 65, 1–6. Sharma, A.K., Venkatesan, G., Mathesh, K., Ram, H., Ramakrishnan, M.A. and Pandey, A.B. 2016. Occurrence and identification of contagious ecthyma in blackbuck. Virus Dis. 27(2), 198–202. Spyrou, V. and Valiakos, G. 2015. Orf virus infection in sheep or goats. Vet. Microbiol. 181, 178–182. Torfason, E.G. and Guðnadóttir, S. 2002. Polymerase chain reaction for laboratory diagnosis of orf virus infections. J. Clin. Virol. 24(2), 79–84. Veraldi, S., Esposito, L., Pontini, P., Vaira, F. and Nazzaro, G. 2019. Feast of sacrifice and Orf, Milan, Italy, 2015–2018. Emerg. Infect. Dis. 25(8), 1585–1586. Windsor, P.A., Nampanya, S., Tagger, A., Keonam, K., Gerasimova, M., Putthana, V., Bush, R.D. and Khounsy, S. 2017. Is orf infection a risk to expanding goat production in developing countries? A study from Lao PDR. Small Rumin. Res. 154(9), 123–128. Yang, H., Meng, Q., Qiao, J., Peng, Y., Xie, K., Liu, Y., Zhao, H., Cai, X. and Chen, C. 2014. Detection of genetic variations in orf virus isolates epidemic in Xinjiang China. J. Basic Microbil. 54(11), 1273–1278. Yogisharadhya, R., Bhanuprakash, V., Venkatesan, G., Balamurugan, V., Pandey, A.B., Shivachandra, S.B. 2012. Comparative sequence analysis of poxvirus A32 gene encoded ATPase protein and carboxyl terminal heterogeneity of Indian orf viruses. Vet. Microbiol. 156(1–2), 72–80. Yusuf, A.O., Mlambo, V., Iposu, S.O. 2018. A nutritional and economic evaluation of Moringa oleifera leaf meal as a dietary supplement in West African Dwarf goats. S. Afr. J. Anim. Sci. 48(1), 81–87. | ||
| How to Cite this Article |
| Pubmed Style Adedeji AJ, Adole JA, Asala OO, Gamawa AAA, Maurice NA, Jambol A, Bolajoko MB, Chima NC, Ifende VI, Wungak YS, Woma TY, Luka PD. A survey of contagious ecthyma and molecular characterization of Orf virus in sheep and goats in Nigeria (2014-2016). Open Vet. J.. 2022; 12(4): 551-561. doi:10.5455/OVJ.2022.v12.i4.18 Web Style Adedeji AJ, Adole JA, Asala OO, Gamawa AAA, Maurice NA, Jambol A, Bolajoko MB, Chima NC, Ifende VI, Wungak YS, Woma TY, Luka PD. A survey of contagious ecthyma and molecular characterization of Orf virus in sheep and goats in Nigeria (2014-2016). https://www.openveterinaryjournal.com/?mno=128469 [Access: January 25, 2026]. doi:10.5455/OVJ.2022.v12.i4.18 AMA (American Medical Association) Style Adedeji AJ, Adole JA, Asala OO, Gamawa AAA, Maurice NA, Jambol A, Bolajoko MB, Chima NC, Ifende VI, Wungak YS, Woma TY, Luka PD. A survey of contagious ecthyma and molecular characterization of Orf virus in sheep and goats in Nigeria (2014-2016). Open Vet. J.. 2022; 12(4): 551-561. doi:10.5455/OVJ.2022.v12.i4.18 Vancouver/ICMJE Style Adedeji AJ, Adole JA, Asala OO, Gamawa AAA, Maurice NA, Jambol A, Bolajoko MB, Chima NC, Ifende VI, Wungak YS, Woma TY, Luka PD. A survey of contagious ecthyma and molecular characterization of Orf virus in sheep and goats in Nigeria (2014-2016). Open Vet. J.. (2022), [cited January 25, 2026]; 12(4): 551-561. doi:10.5455/OVJ.2022.v12.i4.18 Harvard Style Adedeji, A. J., Adole, . J. A., Asala, . O. O., Gamawa, . A. A. A., Maurice, . N. A., Jambol, . A., Bolajoko, . M. B., Chima, . N. C., Ifende, . V. I., Wungak, . Y. S., Woma, . T. Y. & Luka, . P. D. (2022) A survey of contagious ecthyma and molecular characterization of Orf virus in sheep and goats in Nigeria (2014-2016). Open Vet. J., 12 (4), 551-561. doi:10.5455/OVJ.2022.v12.i4.18 Turabian Style Adedeji, Adeyinka Jeremy, Jolly Amoche Adole, Olayinka Oluwafemi Asala, Ahmed Abdulkadir Abdulkadir Gamawa, Nanven Abraham Maurice, Anvou Jambol, Mohammed Bashir Bolajoko, Nneka Chineze Chima, Victoria Isioma Ifende, Yiltawe Simwal Wungak, Timothy Yusufu Woma, and Pam Dachung Luka. 2022. A survey of contagious ecthyma and molecular characterization of Orf virus in sheep and goats in Nigeria (2014-2016). Open Veterinary Journal, 12 (4), 551-561. doi:10.5455/OVJ.2022.v12.i4.18 Chicago Style Adedeji, Adeyinka Jeremy, Jolly Amoche Adole, Olayinka Oluwafemi Asala, Ahmed Abdulkadir Abdulkadir Gamawa, Nanven Abraham Maurice, Anvou Jambol, Mohammed Bashir Bolajoko, Nneka Chineze Chima, Victoria Isioma Ifende, Yiltawe Simwal Wungak, Timothy Yusufu Woma, and Pam Dachung Luka. "A survey of contagious ecthyma and molecular characterization of Orf virus in sheep and goats in Nigeria (2014-2016)." Open Veterinary Journal 12 (2022), 551-561. doi:10.5455/OVJ.2022.v12.i4.18 MLA (The Modern Language Association) Style Adedeji, Adeyinka Jeremy, Jolly Amoche Adole, Olayinka Oluwafemi Asala, Ahmed Abdulkadir Abdulkadir Gamawa, Nanven Abraham Maurice, Anvou Jambol, Mohammed Bashir Bolajoko, Nneka Chineze Chima, Victoria Isioma Ifende, Yiltawe Simwal Wungak, Timothy Yusufu Woma, and Pam Dachung Luka. "A survey of contagious ecthyma and molecular characterization of Orf virus in sheep and goats in Nigeria (2014-2016)." Open Veterinary Journal 12.4 (2022), 551-561. Print. doi:10.5455/OVJ.2022.v12.i4.18 APA (American Psychological Association) Style Adedeji, A. J., Adole, . J. A., Asala, . O. O., Gamawa, . A. A. A., Maurice, . N. A., Jambol, . A., Bolajoko, . M. B., Chima, . N. C., Ifende, . V. I., Wungak, . Y. S., Woma, . T. Y. & Luka, . P. D. (2022) A survey of contagious ecthyma and molecular characterization of Orf virus in sheep and goats in Nigeria (2014-2016). Open Veterinary Journal, 12 (4), 551-561. doi:10.5455/OVJ.2022.v12.i4.18 |