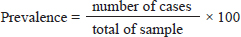| Research Article | ||
Open Vet. J.. 2024; 14(10): 2687-2692 Open Veterinary Journal, (2024), Vol. 14(10): 2687–2692 Research Article Molecular detection of Cryptosporidium spp. among wild rats in Surabaya, East Java, IndonesiaEndang Suprihati1*, Heni Puspitasari2, Elly Nur Indasari2,3, Ponasari Galuh4, Lucia Tri suwanti1,2, Mufasirin Mufasirin1,2, Poedji Hastutiek1 and Boedi Setiawan41Division of Parasitology, Faculty of Veterinary Medicine, Universitas Airlangga, Surabaya, Indonesia 2Toxoplasma Study Group, Institute of Tropical Disease, Universitas Airlangga, Surabaya, Indonesia 3Department of Veterinary Public Health, Faculty of Veterinary Medicine, Universitas Brawijaya, Malang, Indonesia 4Division of Clinic Veteriner, Faculty of Veterinary Medicine, Universitas Airlangga, Surabaya, Indonesia *Corresponding Author: Endang Suprihati. Division of Parasitology, Faculty of Veterinary Medicine, Universitas Airlangga, Surabaya, Indonesia. Email: esuprihati [at] yahoo.co.id Submitted: 26/06/2024 Accepted: 06/09/2024 Published: 31/10/2024 © 2024 Open Veterinary Journal
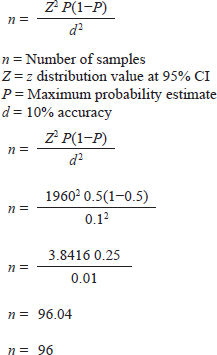
AbstractBackground: Humans and animals who have an acute case of diarrhea can be infected with Cryptosporidium spp. Within the category of water-borne disease, it is a zoonotic disease. The zoonotic disease Cryptosporidium is among the several pathogens carried by wild rats (Rattus spp.). The risk of spreading this disease is rather significant in urban environments because rats are often close to people. Aim: This study aims to detect Cryptosporidium spp. infection in wild rats in Surabaya, East Java, Indonesia. Methods: Through necropsy, a total of 100 wild rats' intestines were sampled for feces. Microscopic observation of the presence of Cryptosporidium was carried out using the float test with a combination of Ziehl Neelsen (ZN) staining. Molecular detection of Cryptosporidium spp. positive results used the Cryptosporidium oocyst wall protein (COWP) gene with polimerase chain reaction method. Results: The results showed that 69 samples were positive for containing Cryptosporidium spp. oocysts and with ZN staining to confirm the diagnosis, the staining results showed Cryptosporidium spp. oocysts dark pink with a clear cavity inside with a percentage of 95.65% in Rattus norvegicus and 61.03% in Rattus tanezumi. In residential and densely populated environments the percentage of Cryptosporidium spp. amounted to 66.66% and in the market environment amounted to 74.19%. The percentage of Cryptosporidium spp. in the North Surabaya region was 42.85%, South Surabaya 100%, West Surabaya 37.5%, East Surabaya 81.39%, and Central Surabaya 65.38%. Molecular detection of Cryptosporidium spp. positive results were obtained using the COWP gene 550 bp. Conclusion: This study aims to detect Cryptosporidium spp. infection in wild rats in Surabaya, East Java, Indonesia. The high number of cases of cryptosporidiosis in wild rats has the potential to be a reservoir for the spread of the disease. The Cryptosporidium spp can detected with COWP in 550 bp in wild rats in Surabaya, East Java, Indonesia. Keywords: COWP gene, Cryptosporidium spp., Wild rat, Ziehl-Neelsen, Zoonosis. IntroductionCryptosporidiosis is a zoonotic disease and is classified as a waterborne disease, that is transmitted between living creatures due to contamination in the form of microorganisms or dangerous substances through water (Mufasirin et al ., 2020). More than 40 species and genotypes of Cryptosporidium have been identified worldwide (Zahedi et al., 2021). In rodents, a total of 21 species and 21 genotypes of Cryptosporidium were found (Zhao et al., 2019). The Cryptosporidium species most frequently detected in wild rats are Cryptosporidium parvum and Cryptosporidium muris (Koehler et al., 2018). The infective form of Cryptosporidium spp. are oocysts excreted in the feces of infected hospes. The clinical signs and symptoms of cryptosporidiosis are contingent upon the immunological condition of the individual. In immunocompetent individuals, infection with Cryptosporidium spp. can be asymptomatic or with symptoms of acute diarrhea that can heal itself. Meanwhile, in immunocompromised individuals, diarrhea can become chronic and can even cause death (Roy et al., 2004). The prevalence of Cryptosporidium spp. in animals varies greatly, in rodents as much as 22% in Europe (Wijayanti, 2017), France in 2021 at 2.1%–63.0% (Livia et al., 2021), China at 8.2% (Zhao et al., 2015), and Japan at 8% using the immunofluorescence assay (IFA) technique and 38% positive with the Nested polimerase chain reaction (PCR) technique (Kimura et al., 2007). This research aims to detect Cryptosporidium infection in wild rats in Surabaya parasitologically and molecularly. The purpose of this study is to identify whether wild rats harbor the protozoan Cryptosporidium spp. The broad range and high incidence of Cryptosporidium spp. in wild rodents contribute significantly to the spread of the disease to humans and have the potential to become a significant source of contaminated water. Materials and MethodsSampling techniqueThe study was carried out in 2023 from June to October. Surabaya, East Java, Indonesia, was the site of the sample. The samples were examined under a microscope at Universitas Airlangga's Faculty of Veterinary Medicine's Laboratory of Veterinary Parasitology. The Institute of Tropical Disease at Universitas Airlangga carried out PCR testing. Under Airlangga University's 2.KEH.080.07.2022, the Animal Care and Use Committee of the Faculty of Veterinary Medicine authorized the experimental procedure. Wild rat samples are collected in five Surabaya areas (North, West, South, East, and Central of Surabaya) as well as in each area where sampling is done including the communities, the market environment, and the densely populated housing environment habitation. The Lemeshow formula was utilized to calculate the sample in this investigation (Lemeshow et al., 1990) in a state of population unknown. The following computations were made:
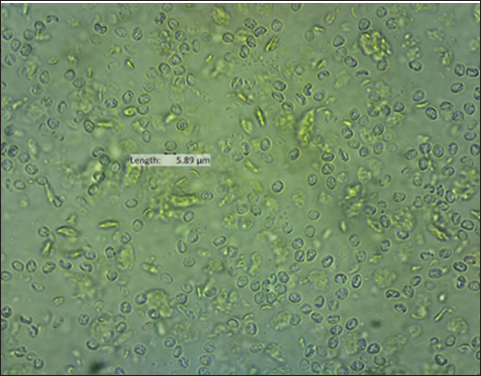
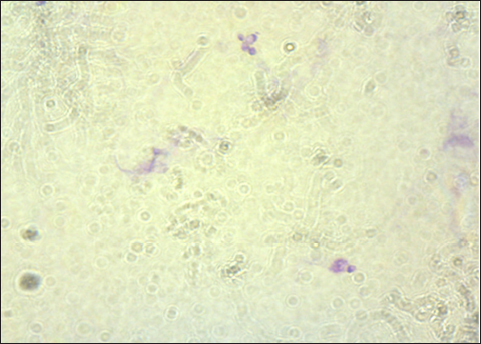
The result of the calculation of the formula Lemeshow obtained the sample number result. The minimum required in this study is 96 samples, the total number of samples. In this study, there were 100 wild rats. Parasitological and Ziehl Neelsen (ZN) methodFecal samples were taken from the digestive tract of rats by necropsy, then the feces were examined using the sugar flotation centrifugation method. ZN staining technique was applied on smears. After the smear has dried and been fixed with a Bunsen burner, carbol fushin is applied to the slide and heated until it turns to steam. The slide is then rinsed with water and, depending on the smear's thickness, decolored for 1 minute with 2.5% sulfuric acid. The slide is then stained once more by dripping 1% methylene blue for 1 minute, dried, and examined at 400× or 1,000× magnification (Tahvildar and Salehi, 2014). Polymerase chain reactionExtraction by sonication process first and followed by the Qiagen® kit. The primer design uses the Cryptosporidium oocyst wall protein gene (COWP) which is found in all species of Cryptosporidium spp. COWP primers: forward primer 5′-GTAGATAATGGAAGAGATTGTG-3′ and reverse primer 5′-GGACTGAAATACAGGCATTATC TTG-3′ with 550 bp. The steps involved in PCR amplification are as follows: 3 minutes of initial denaturation at 94°C; 30 seconds of denaturation at 94°C, 30 seconds of annealing at 55°C, 60 seconds of extension at 72°C, and 7 minutes of elongation at 72°C. Electrophoresis is performed after PCR findings (Spano et al., 1997). Ethical approvalThe Animal Care and Use Committee of the Faculty of Veterinary Medicine at Airlangga University, with approval number 2.KEH.080.07.2022, authorized the experimental procedure. ResultsParasitological testMicroscopic examination of rat feces samples in Surabaya using the floating method aimed at detecting Cryptosporidium spp. oocysts. The results showed that 69 out of 100 samples contained Cryptosporidium spp. oocysts. The oocysts found were round in shape with a diameter of 3.67 μm (Fig. 1). This is in accordance with the report by Mufasirin et al. (2020) in their research that the size of the oocysts of Cryptosporidium spp. 2–6 μm and has a round shape with four sporoozytes inside. ZN methodFeces examination techniques can be combined with ZN staining to confirm the diagnosis, the staining results show Cryptosporidium spp oocysts. round, dark pink in color with a clear cavity inside (Fig. 2). This is in accordance with the report by Jassim and Al-Mussawi (2021) in their research that the oocysts of Cryptosporidium spp. will be pink in color and is in accordance with the research of Rekha et al. (2016) that with ZN staining of Cryptosporidium spp oocysts. It appears dark pink in color with a clear cavity inside.
Fig. 1. Oocysts of Cryptosporidium spp.
Fig. 2. Oocysts of Cryptosporidium spp. with ZN staining. Prevalence Cryptosporidium spp of wild rats in SurabayaThe prevalence in parasitological tests is the percentage of rats infected with Cryptosporidium spp. compared to the total of sample. The prevalence rate is calculated using:
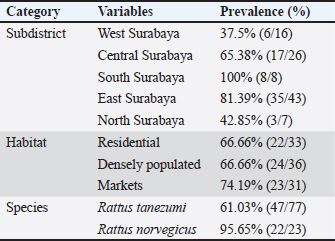
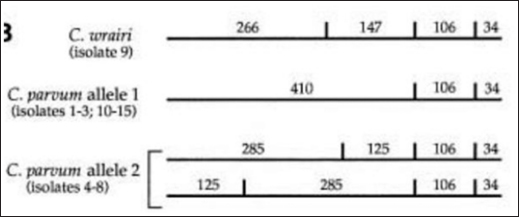
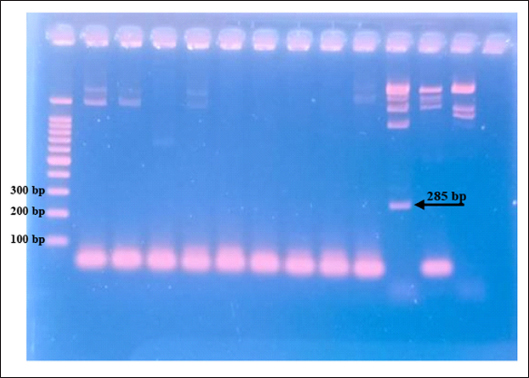
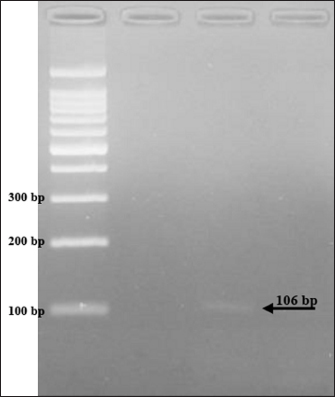
The results showed that the prevalence of Cryptosporidium spp. oocysts were 69% (69/100). Prevalence of Cryptosporidium spp. oocysts in Rattus norvegicus were 95.65% (22/23) and 61.03% (47/77) in Rattus tanezumi. Prevalence of Cryptosporidium spp. oocysts in wild rats in residential were 66.66% (22/33), in densely populated environments, were 66.66% (24/36), and in the market environment 74.19% (23/31). Prevalence of Cryptosporidium spp. in wild rats in the North Surabaya region, it was 42.85% (3/7), South Surabaya 100% (8/8), West Surabaya 37.5% (6/16), East Surabaya 81.39% (35/43), and Central Surabaya 65.38% (17/26) (Table 1). Molecular detection of Cryptosporidium spp. positive results were obtained using the COWP gene with PCR method. Molecular detection of Cryptosporidium spp. in wild ratsData from the results of microscopic tests and molecular tests were analyzed qualitatively and explained descriptively. Molecular analysis by a number of amplified band fragments is based on research by Spano et al. (1997), there are 34, 106, 125, and 285 bp (Fig. 3). Molecular testing with PCR aims to confirm positive results for Cryptosporidium spp. after microscopic examination. Detection of Cryptosporidium spp. in this study used the COWP gene with a band of 550 bp with fragments of 34, 106, 125, and 285 bp (Spano et al., 1997). The results of the research show visualization of the PCR product using the COWP gene, amplification of a DNA band of 285 bp with an annealing temperature of 52.6°C with the DNA preextraction method using the heating method, and amplification of a DNA band of 106 bp temperature annealing 48°C DNA preextraction method using the sonication. PCR results can be seen in Figures 4 and 5. Table 1. Demographic data for prevalence toxoplasmosis of wild rats in Surabaya.
The results detection of Cryptosporidium spp. on wild rats in Surabaya using the PCR method, positive results were obtained 285 bp was amplified with an annealing temperature at 52.6°C and 106 bp with an annealing temperature at 48°C using the COWP gene primer. The results obtained are only two bands appeared out of the four bands that should appear in the PCR product; 34, 106, 125, and 285 bp (Spano et al., 1997). DiscussionDetection of Cryptosporidium spp. in this study used the COWP gene with a band of 550 bp with fragments of 34, 106, 125, and 285 bp (Spano et al., 1997). The only known apicomplexan oocyst membrane protein is the Cryptosporidium oocyst wall protein 1 (COWP1) gene, which codes for the COWP. COWP1 is found in the wall-forming bodies of mature macrogametes as well as the inner oocyst wall, according to immunoelectron microscopy results (Templeton et al., 2004). The COWP gene PCR has a specificity value of 99.6% and a sensitivity value of 90% (Weinreich et al., 2021). Cryptosporidium is one of the numerous zoonotic infections carried by wild rats (Rattus spp.). There is a significant chance that this disease will spread because rats and people live close together in an urban environment (Zhao et al., 2019). One possible source of infection is rodents. There may also be mechanical transmission by insects, birds, and people (Martin and Aitken, 2000). Season and the quantity of rats in the community are additional risk variables. High levels of environmental contamination are ensured by the enormous number of oocysts released upon infection (Dixon et al., 2011).
Fig. 3. Length of amplified band fragments (Spano et al., 1997).
Fig. 4. PCR results of the COWP gene 285 bp.
Fig. 5. PCR results of the COWP gene 106 bp. When the infectious oocyst is consumed by the new host, infection results. The upper gastrointestinal tract becomes excited after consumption. When oocysts enter the digestive system, they produce infectious sporozoites, or trophozoites, which cling to intestinal epithelial cells (Bouzid et al., 2018). In a host that is vulnerable, one oocyst is enough to cause infection and illness. The fecal-oral pathway is the means by which environmentally resistant oocysts spread (Smith et al., 2007). Because of the high likelihood that Cryptosporidium oocysts in soil may find their way into water supplies, populations residing close to rivers should be viewed as possible sources of waterborne disease (Ghazy et al., 2015). AcknowledgmentsThe Universitas Airlangga Faculty of Veterinary Medicine is greatly appreciated by the authors for their support of this research. Conflict of interestThere is no conflict of interest, according to the authors. FundingThe authors are grateful to Airlangga University for providing contract number 254/UN3/2023 for the Faculty's Excellence Research Scheme. Authors’ contributionsES coordinated the research, authored the article manuscripts, reviewed the literature, and designed and conceptualized the proposal. PH, BS, PG MM, HP, ENI, PH Rearing and trapping wild rats, gathering information, reviewing the literature, and analyzing the information; SK also worked with flow cytometry and data processing. Each author edited and proofread the work. Data availabilityThe manuscript contains all the data needed to support the study's conclusions; no other data sources are needed. ReferencesBouzid, M., Kintz, E. and Hunter, P. 2018. Risk factors for Cryptosporidium infection in low and middle income countries: a systematic review and meta-analysis. PLoS Neglected Trop. Dis. 12(6), 1–13. Dixon, B., Parrington, L., Cook, A., Pintar, K., Pollari, F., Kelton, D. and Farber, J. 2011. The potential for zoonotic transmission of Giardia duodenalis and Cryptosporidium spp. from beef and dairy cattle in Ontario, Canada. Vet. Parasitol. 175(1–2), 6–20. Ghazy, A., Abdel S. and Shaapan, R. 2015. Cryptosporidiosis in animals and man: 1. Taxonomic classification, life cycle, epidemiology and zoonotic importance. Asian J. Epidemiol. 8, 48–63. Jassim, A.H. and Al-Mussawi, K.A.M. 2021. Comparison of Three Diagnostic Methods for Detecting the Cryptosporidium Parvum Parasite in Karbala Governorate / Iraq. Annal. Rom. Soc. Cell Biol. 25(4), 13966–13972. Kimura, A., Edagawa A., Okada K., Takimoto A., Yonesho S. and Karanis, P. 2007. Detection and genotyping of Cryptosporidium from brown rats (Rattus norvegicus) captured in an urban area of Japan. Parasitol. Res. 100, 1417–1420. Koehler, A., Wang, T., Haydon, S., and Gasser, R. 2018. Cryptosporidium viatorum from the native Australian swamp rat Rattus lutreolus—an emerging zoonotic pathogen? Int. J. Parasitol. Parasites. Wildl. 7, 18–26. Lemeshow, S., Hosmer, W., Klar J. and Lwanga, K. 1990. Adequacy of sample size in health studies. England, UK: John Wiley & Sons Ltd., pp: 1–2. Livia, G., Fernández, Á., Feliu, C., Miquel, J., Quilichini, Y. and Foronda P. 2021. Cryptosporidium spp. in wild murids (Rodentia) from Corsica, France. Parasitol. Res. 121, 345–354. Martin, W. and Aitken, I., 2000. Diseases of sheep, 3rd ed.. Oxford, UK: Blackwell Science Ltd., pp: 153–159. Mufasirin, R., Nunuk, D., Fedik, A., Lucia, T., Endang, S., Didik, H. and dan Mufasirin. 2020. Deteksi Cryptosporidium canis pada Anjing di Kota Surabaya. Vet. J. 21(2), 176–182. Rekha, K.M., Puttalakshmamma, G.C. and D'Souza, P.E. 2016. Comparison of different diagnostic techniques for the detection of cryptosporidiosis in bovines. Vet. World. 9(2), 211–215. Roy, S., DeLong, S., Stenzel, S., Shiferaw, B., Roberts, J., Khalakdina, A., Marcus, R., Segler, S., Shah, D., Thomas, S., Vugia, D., Zansky, S., Dietz, V. and Beach, M. 2004. Risk factor for sporadic cryptosporidiosis among immunocompetent persons in the United States from 1999 to 2001. J. Clin. Microbiol. 42(7), 2944–2951. Smith, H., Caccio, S., Cook, N., Nichols, R. and Tait, A. 2007. Cryptosporidium and Giardia as foodborne zoonoses. Vet. Parasitol. 149, 29–40. Spano, F., Putignani, L., McLauchlin, J., Casemore, D. and Crisanti, A. 1997. PCRRFLP analysis of the Cryptosporidium oocyst wall protein (COWP) gene discriminates between C. wrairi and C. parvum, and between C. parvum isolates of human and animal origin. FEMS Microbiol. Lett. 150(2), 209–217. Tahvildar, B. and Salehi, N. 2014. Detection of Cryptosporidium infection by modified ziehl-neelsen and PCR methods in children with diarrheal samples in pediatric hospitals in Tehran. Gastroenterol. Hepatol. Bed Bench. Spring. 7(2), 125–130. Templeton, T., Lancto, J., Vigdorovich, C., Liu, V., London, C., Hadsall, K. and Abrahamsen, M. 2004. The Cryptosporidium oocyst wall protein is a member of a multigene family and has a homolog in Toxoplasma. Infect. Immun. 72(2), 980–987. Weinreich, F., Hahn, A., Eberhardt, K.A., Feldt, T., Sarfo F.S., Di Cristanziano, V., Frickmann, H. and Loderstädt, U. 2021. Comparison of three real-time PCR assays targeting the SSU rRNA gene, the COWP gene and the DnaJ-Like protein gene for the diagnosis of Cryptosporidium spp. in stool samples. Pathogens (Basel, Switzerland). 10(9), 1131. Wijayanti, T. 2017. Cryptosporidiosis in Indonesia. Balaba. 13(01), 73–82. Zahedi, A., Bolland, S., Oskam, C., and Ryan, U. 2021. Cryptosporidium abrahamseni n. sp. (Apicomplexa: Cryptosporidiiae) from red-eye tetra (Moenkhausia sanctaefilomenae). Exper. Parasitol. 223, 108089. Zhao, Z., Wang, R., Zhao, W., Qi, M., Zhao, J., Zhang, L., Li, J. and Liu, A. 2015. Genotyping and subtyping of Giardia and Cryptosporidium isolates from commensal rodents in China. Cambridge University Press, vol. 142, no. 6, pp: 800–806. Zhao, W., Zhou, H., Huang, Y., Xu, L., Rao, L., Wang, S., Wang, W., Yi, Y., Zhou, X., Wu, Y., Ma, T., Wang, G., Hu, X., Peng, R., Yin, F. and Lu, G. 2019. Cryptosporidium spp. in wild rats (Rattus spp.) from the Hainan Province, China: molecular detection, species/genotype identification and implications for public health. Int. J. Parasitol. Parasites Wildl. 9, 317–321. | ||
| How to Cite this Article |
| Pubmed Style Suprihati E, Puspitasari H, Indasari EN, Galuh P, Suwanti LT, Mufasirin M, Hastutiek P, Setiawan B. Molecular detection of Cryptosporidium spp. among wild rats in Surabaya, East Java, Indonesia. Open Vet. J.. 2024; 14(10): 2687-2692. doi:10.5455/OVJ.2024.v14.i10.18 Web Style Suprihati E, Puspitasari H, Indasari EN, Galuh P, Suwanti LT, Mufasirin M, Hastutiek P, Setiawan B. Molecular detection of Cryptosporidium spp. among wild rats in Surabaya, East Java, Indonesia. https://www.openveterinaryjournal.com/?mno=202065 [Access: January 25, 2026]. doi:10.5455/OVJ.2024.v14.i10.18 AMA (American Medical Association) Style Suprihati E, Puspitasari H, Indasari EN, Galuh P, Suwanti LT, Mufasirin M, Hastutiek P, Setiawan B. Molecular detection of Cryptosporidium spp. among wild rats in Surabaya, East Java, Indonesia. Open Vet. J.. 2024; 14(10): 2687-2692. doi:10.5455/OVJ.2024.v14.i10.18 Vancouver/ICMJE Style Suprihati E, Puspitasari H, Indasari EN, Galuh P, Suwanti LT, Mufasirin M, Hastutiek P, Setiawan B. Molecular detection of Cryptosporidium spp. among wild rats in Surabaya, East Java, Indonesia. Open Vet. J.. (2024), [cited January 25, 2026]; 14(10): 2687-2692. doi:10.5455/OVJ.2024.v14.i10.18 Harvard Style Suprihati, E., Puspitasari, . H., Indasari, . E. N., Galuh, . P., Suwanti, . L. T., Mufasirin, . M., Hastutiek, . P. & Setiawan, . B. (2024) Molecular detection of Cryptosporidium spp. among wild rats in Surabaya, East Java, Indonesia. Open Vet. J., 14 (10), 2687-2692. doi:10.5455/OVJ.2024.v14.i10.18 Turabian Style Suprihati, Endang, Heni Puspitasari, Elly Nur Indasari, Ponasari Galuh, Lucia Tri Suwanti, Mufasirin Mufasirin, Poedji Hastutiek, and Boedi Setiawan. 2024. Molecular detection of Cryptosporidium spp. among wild rats in Surabaya, East Java, Indonesia. Open Veterinary Journal, 14 (10), 2687-2692. doi:10.5455/OVJ.2024.v14.i10.18 Chicago Style Suprihati, Endang, Heni Puspitasari, Elly Nur Indasari, Ponasari Galuh, Lucia Tri Suwanti, Mufasirin Mufasirin, Poedji Hastutiek, and Boedi Setiawan. "Molecular detection of Cryptosporidium spp. among wild rats in Surabaya, East Java, Indonesia." Open Veterinary Journal 14 (2024), 2687-2692. doi:10.5455/OVJ.2024.v14.i10.18 MLA (The Modern Language Association) Style Suprihati, Endang, Heni Puspitasari, Elly Nur Indasari, Ponasari Galuh, Lucia Tri Suwanti, Mufasirin Mufasirin, Poedji Hastutiek, and Boedi Setiawan. "Molecular detection of Cryptosporidium spp. among wild rats in Surabaya, East Java, Indonesia." Open Veterinary Journal 14.10 (2024), 2687-2692. Print. doi:10.5455/OVJ.2024.v14.i10.18 APA (American Psychological Association) Style Suprihati, E., Puspitasari, . H., Indasari, . E. N., Galuh, . P., Suwanti, . L. T., Mufasirin, . M., Hastutiek, . P. & Setiawan, . B. (2024) Molecular detection of Cryptosporidium spp. among wild rats in Surabaya, East Java, Indonesia. Open Veterinary Journal, 14 (10), 2687-2692. doi:10.5455/OVJ.2024.v14.i10.18 |