| Original Article | ||
Open Vet. J.. 2021; 11(4): 581-586 Open Veterinary Journal, (2021), Vol. 11(4): 581–586 Original Research Efficacy of two primer sets used in the sex identification of rufous-winged buzzard (Butastur liventer)Paitoon Kaewhom1 and Kanokrat Srikijkasemwat2*1Faculty of Agricultural Technology, Burapha University, Sa Kaeo Campus, Sa Kaeo, Thailand 2Department of Animal Production Technology and Fisheries, Faculty of Agricultural Technology, King Mongkut’s Institute of Technology Ladkrabang, Bangkok, Thailand *Corresponding Author: Kanokrat Srikijkasemwat. Faculty of Agricultural Technology, King Mongkut’s Institute of Technology Ladkrabang, Bangkok, Thailand. Email: kanokrat.sr [at] kmitl.ac.th Submitted: 27/06/2021 Accepted: 09/10/2021 Published: 27/10/2021 © 2021 Open Veterinary Journal
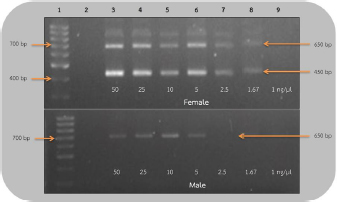
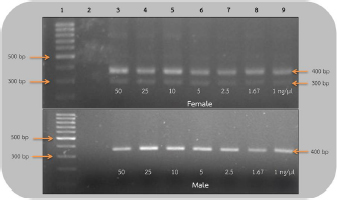
AbstractBackground: The rufous-winged buzzard (Butastur liventer) belongs to the order Accipitriformes, which is monomorphic, resulting in the difficulty to identify the gender. However, sex determination is important for predicting the sex ratio of this buzzard in nature in order to avoid its extinction. Aim: We aimed to develop a primer set that is able to sex the rufous-winged buzzard through polymerase chain reaction (PCR) amplification and compare the efficacy of the two sets of primers by using PCR technique. Methods: In the following, sensitivity refers to the smallest DNA concentration that allowed us to accurately sex a bird and specificity refers to the ability to clearly distinguish the sex based on the visual appearance of the bands. Blood samples were collected from captive buzzards. The DNA was extracted from them and was diluted to 50, 25, 10, 5, 2.5, 1.67, and 1 ng/μl. Two sets of primers, including P2/NP/MP and 2550F/2718R, were used to amplify the chromo-helicase DNA binding (CHD) gene of known gender buzzards using the PCR process to determine gender and to compare their sensitivity. To measure specificity, both primers were used to amplify CHD gene fragment of other unknown gender birds. Results: The lowest concentration of the DNA template where P2/NP/MP could amplify DNA fragments was 1 ng/μl, and this set of primers could identify the gender of all birds correctly, giving 100% specificity. On the other hand, the 2550F/2718R could amplify the DNA fragments from 5 ng/μl, and it had only 78% specificity. Conclusion: The P2/NP/MP primer set was able to correctly identify the gender of rufous-winged buzzard through PCR amplification with high specificity and sensitivity. Keywords: CHD gene, PCR technique, Gender identification, Birds. IntroductionThe rufous-winged buzzard (Butastur liventer), a homeothermic species, belongs to the order Accipitriformes and class Aves. It is a bird of prey found in Southeast Asia and Southern China. It has strong feet, sharp talons, and a powerful well-developed beak for tearing the prey. Moreover, it has keen vision, excellent hearing, and a superior flying ability. It hunts during the day or in the twilight. In Thailand, Accipitriformes are protected. Additionally, since they are at the very top of the food chain, they play a crucial role in maintaining a stable ecosystem. It is permitted to own an Accipitriformes, but the breeder must be approved by the Director General of the Department of National Parks, Wildlife and Plant Conservation. Aves generally have different characteristics between genders, but some species, particularly birds of prey, are monomorphic, i.e., both genders have similar characteristics and cannot be simply distinguished. It is particularly difficult to identify the gender of young birds, as both genders are almost identical in appearance. There are several techniques for determining gender, e.g., observing appearance and behavior, anatomical measurements, laparoscopy, determining the level of steroid hormones in the plasma or feces, and chromosome identification. These techniques are time-consuming and expensive, and may also injure the vital organs of birds. These techniques require specialists or it could lead to wrong classification. Molecular techniques are more accurate and effective, e.g., microsatellite, minisatellite, single strand conformation polymorphism, random amplification of polymorphic DNA, amplification fragment length polymorphism, restriction fragment length polymorphism, amplification refractory mutation system (ARMS), and triple-primer polymerase chain reaction (PCR). Many of these are based on the chromo-helicase DNA binding (CHD) gene, which encodes the CHD protein 1, which is present in both gender chromosomes, W and Z, and the helicase domain of this gene is highly conserved in other species of this class. Birds can be sexed by PCR using the primers that are specific to CHD. The W chromosome in the CHD gene is slightly longer than the Z one because of additional bases in the intron region. Furthermore, the intron lengths for chromo-helicase DNA blinding protein W-linked (CHDW) and chromo-helicase DNA blinding protein Z-linked (CHDZ) vary in different species of birds. Thus, multiple primers were designed to determine the various intron lengths in CHD genes. However, primers are specific to a particular species. Therefore, we aimed to find primers that are specific to the sex chromosomes of the rufous-winged buzzard and compare the efficacy of two sets of primers using the PCR technique. Materials and MethodsCollection of blood samplesThirteen captive buzzards’ blood samples were collected by puncturing the wing vein. Four of these were collected from two pairs, whose genders were known from pairing and producing offspring. The other nine samples were collected from birds of unknown gender. Then, the samples were kept in tubes, coated with EDTA to prevent blood clotting, at −20°C, until DNA could be extracted. DNA extraction and dilutionDNA was extracted from the blood samples by acid phenol extraction (Chomczynsky and Sacchi, 1987); then the DNA concentration and its quality (ratio A260/A280) were measured by a spectrophotometer (Thermo Scientific™ NanoDrop™, USA). All samples were then diluted to 50 ng/μl. Serial dilution used the ratios 1:1, 1:5, 1:10, 1:20, 1:30, and 1:50. Therefore, the DNA of the four known gender birds was diluted to 25, 10, 5, 2.5, 1.67, and 1 ng/μl. These were DNA templates which were used in DNA amplification. DNA amplificationThe DNA templates of 50, 25, 10, 5, 2.5, 1.67, and 1 ng/μl samples were amplified. Two primer sets, which are specific to the CHD gene P2/NP/MP (Ito et al., 2003) and 2550F/2718R (Fridolfsson and Ellegren, 1999) (Table 1) were used. The PCR amplification of 2550F/2718R primer pair used a volume of 10 μl. The final reaction conditions were 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 20 mM dNTPs, 200 mM MgCl2, 1 pM of sense and anti-sense primers, 5.0 units DNA polymerase (Invitrogen®, USA), and the DNA templates of the listed concentrations. The PCR used a Primus 96 plus thermocycler. Firstly, the DNA templates were denatured (94°C, 5 minutes); PCR was carried out for 35 cycles with each cycle including template DNA denaturation (94°C, 1 minute), annealing (50°C, 1 minute), and an extension step (72°C, 30 seconds). The final extension step (72°C, 5 minutes) ensured full-length polymerization of the target DNA. Similarly, the amplification of P2/NP/MP primer used the same steps, except that the annealing step was at 50°C. Finally, the amplified PCR products were analyzed by electrophoresis in 1.5% (w/v) agarose gel and visualized using ethidium bromide and a UV light. The lowest concentration of DNA templates, which clearly showed the fragments in agarose gel in the previous experiment, was then used to determine the sex of nine unidentified gender birds. The CHD genes were amplified using both sets of primers under the same conditions. Ethical approvalThe owners of the buzzards had the required approvals from the Thai Government Department of National Parks, Wildlife and Plant Conservation to keep them. The Animal Care and Use Committee of KMITL also approved this project. ResultsEvaluation of sensitivity of primers for gender identificationThis experiment used 50 ng/reaction, and it was then diluted to give 25, 10, 5, 2.5 1.67, and 1 ng/μl of the known gender bird blood samples. These DNA templates were amplified to compare the sensitivities of the 2550F/2718R and P2/NP/MP primers. These primers amplified the introns of CHDW and CHDZ genes. Both primers were expected to display one fragment in male and two fragments in female, since the male chromosome has only Z and the female has both W and Z chromosomes. As expected, using the 2550F/2718R primer flanking the intron, females displayed a fragment of 600–700 base pairs and another of 400–500 base pairs, which were produced in the PCR, while males only showed one fragment of 600–700 base pairs. In the PCR products of female birds, the bands were clearly seen when the DNA template concentration was ≥1.67 ng/μl. However, the DNA fragments of male birds were only apparent at ≥5 ng/μl. Hence, this pair of primers could determine the sex of the buzzard at concentration of ≥5 ng/μl (Fig. 1). The P2/NP/MP primer also gave the results as expected, i.e., accurate sex identification of all four known gender birds. Male birds displayed only one fragment of 300–400 base pairs. Meanwhile, females showed two fragments: 300–400 base pairs and 200–300 base pairs. Furthermore, this set of primers was so sensitive that they were able to identify the gender from concentrations ≥1 ng/μl, with clear band patterns (Fig. 2). Table 1. Sequence of the two primer sets compared in the current study.
Fig 1. Analysis of PCR products of the CHD gene using 2550F/2718R primer run on 1.5% agarose gel electrophoresis. 10 μl of the PCR products was loaded onto each lane of agarose gel. Lane 1=DNA marker (100 bp), lane 2=negative control, lanes 3–9=PCR products of female and male blood samples at DNA concentrations of 50, 25, 10, 5, 2.5, 1.67, and 1 ng/μl.
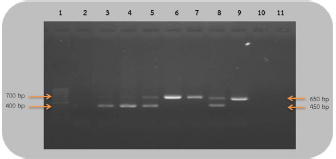
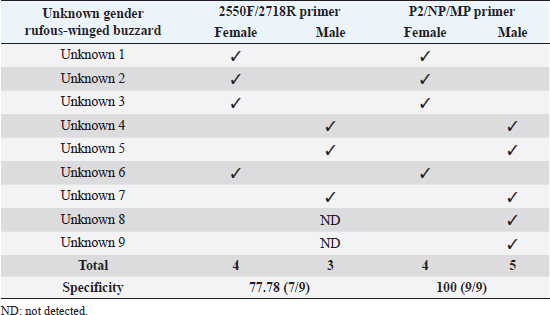
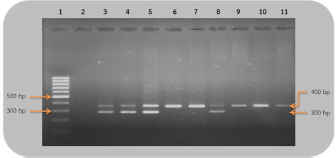
Fig 2. Analysis of PCR products of the CHD gene using the P2/NP/MP primer run on 1.5% agarose gel electrophoresis. 10 μl of PCR mixture was loaded on each lane of agarose gel. Lane 1=DNA marker (100 bp), lane 2=negative control, lane 3–9=PCR products of female and male at DNA concentrations of 50, 25, 10, 5, 2.5, 1.67, and 1 ng/μl. Comparison of specificity of gender identification between the two sets of primersTo identify the sex of the nine unknown gender birds, two sets of primers were used to compare specificity. Since 5 ng/μl was the lowest concentration that showed apparent fragments of PCR products in the previous experiment, this concentration was used in this experiment. Using the 2550F/2718R primer, three birds were identified as male, four as female, and two were unidentified, since the tests displayed no fragments. Thus, the specificity of 2550F/2718R primer was 78% (7/9) (Fig. 3 and Table 2).
Fig 3. Analysis of PCR products of the CHD gene using the 2550F/2718R primer run on 1.5% agarose gel electrophoresis. 10 μl of PCR mixture was loaded on each lane of agarose gel. Lane 1=DNA marker (100 bp), lane 2=negative control, lane 3–11=PCR products of the CHD gene from unknown samples. The P2/NP/MP primer showed that five were male and four were female. Therefore, this set of primers had 100% (9/9) specificity, or higher than the first set (Fig. 4 and Table 2). They also produced clearer band patterns. DiscussionImportant factors that generally affect gender determination include the source of samples, extraction method, DNA concentration, and primers used in PCR process. Firstly, the sources of samples could be blood (Ellegren, 1996; Wang et al., 2007), feathers (Jensen et al., 2003; Sacchi et al., 2004; Costantini et al., 2008), tissue (Kahn et al., 1998; Fridolfsson and Ellegren, 1999; Wang et al., 2007), shell membrane, blood vessels (Jensen et al., 2003), and feces (Robertson et al., 1999; Idaghdour et al., 2003). In this study, the DNA was extracted from the buzzard blood samples because it was convenient to extract DNA and to obtain sufficient DNA for further experiments. However, it was very important to be cautious about collecting blood samples from the birds in order to prevent them from becoming injured or stressed. Secondly, there are several ways to extract DNA: some of these include the phenol–chloroform (Sambrook et al., 1989), the guanidium thiocyanate (Hammond et al., 1996), and acid guanidinium thiocyanate–phenol–chloroform methods (Chomczynsky and Sacchi, 1987). Although they are time-consuming and laborious, and involve toxic chemicals, good DNA yields and quality can be obtained. An alternative approach, a lesser time-consuming method, is Chelex extraction (Walsh et al., 1991; Dubiec and Magdalena, 2006) since there is no digestion step required. Another method that can be used to avoid toxicity is a commercial kit, which is easier to carry out, and is faster and safer. Thirdly, the concentration of DNA also influences bird gender identification. Reaction with a very low DNA concentration of the template produces low yields while reactions with excessive DNA template can be plagued by nonspecific amplification, leading to smearing in the agarose gel. Lastly, primers that are specific to the target gene are particularly important. Previous studies have reported that the DNA concentrations used ranged from 50 to 200 ng/reaction (Ito et al., 2003; Reddy et al., 2007; Lee et al., 2008; Garofalo et al., 2016). The blood samples of the four known gender birds were extracted and diluted to obtain 1–50 ng/μl DNA, and two sets of primers were used in the amplification to compare sensitivity of both primers, a comparison that has not been reported in other studies. This study revealed that only 1 ng/μl DNA template could be amplified by the P2/NP/MP primer, showing clear fragments in the agarose gel, whereas the 2550F/2718R primer could amplify a 5 ng/μl template. It was apparent that high specificity and high sensitivity primers were able to amplify DNA fragments even at low DNA concentrations. Here, we found that the P2/NP/MP primer set was more sensitive and more specific to our buzzard, than 2550F/2718R. Hence, it was possible to collect DNA sample by other noninvasive sampling methods, for example, collecting feathers, egg shell membranes, or feces. Although these give small amounts of DNA, PCR amplification can use the P2/NP/MP primer. This information can be developed to predict the sex ratio of buzzard in a particular natural area to determine whether there is a risk of extinction. Table 2. Sex identification of unknown gender rufous-winged buzzard (n=9).
Fig 4. Analysis of PCR products of the CHD gene using P2/NP/MP primer run on 1.5% agarose gel electrophoresis. 10 μl of PCR mixture was loaded on each lane of agarose gel. Lane 1=DNA marker (100 bp), lane 2=negative control, lane 3–11=PCR products of CHD gene from the unknown samples. The first set of primers used in gender identification was P2/P3 (Griffiths and Tiwari, 1995). It was not commonly used since the amplification of exon in CHDW and CHDZ genes differed by only 60–110 base pairs, depending on the species. After that, more primers were developed to determine the sex of birds, including P2/P8 (Griffiths et al., 1998), 1237L/1272H (Kahn et al., 1998), and 2550F/2718R (Fridolfsson and Ellegren, 1999). The P2/P8 primer was used to determine the sex of Eurasian oystercatchers. Eighty samples were tested. Seventy-five of them were correctly identified, while the CHD genes of five of them were not amplified by this pair of primers. The specificity of the P2/P8 primer was 94% (75/80) (Watson et al., 2004). Another study determined the gender of 80 different bird species, using 1237L/1272H and 2550F/2718R, in the same PCR conditions. It was found that the 1237L/1272H primer identified the gender of 63 species, whereas in only 59 species was the gender correctly determined by the 2550F/2718R primer. Therefore, the specificity of 1237L/1272H was 79% (63/80), whereas for 2550F/2718, it was 74% (59/80) (Wang et al., 2007). These studies, showed that selecting a suitable primer set, which was highly specific to the species of interest, was particularly important. In 2003, Ito et al. (2003) developed the ARMS-PCR (multiplex with P2/NP/MP primer) for vulture gender determination, giving strong signal bands. The NP/MP primer pair amplified the CHDW gene, whereas CHDZ was amplified by P2/NP primer. The PCR products from the CHDZ gene of male birds were 369–375 base pairs (depending on the species), whereas, for females, the PCR products of CHDZ had 378 base pairs compared to CHDW 293 base pairs (Prabhakar et al., 2012). Since both genes differed by approximately 100 base pairs, fragments were effectively separated by agarose gel electrophoresis. Thammakarn et al. (2007) studied the gender identification of 38 Psittacines in six species. By using the P2/NP/MP primer set, the sexes of all 38 birds were correctly identified with 100% (38/38) specificity. There are only a few studies of rufous-winged buzzards, as they are found in Southeast Asia and Southern China. The primers that have been found to identify the gender of other Accipitriformes species were the 2550F/2718R (Fridolfsson and Ellegren, 1999) and P2/NP/MP primers (Ito et al., 2003). We investigated the sensitivity and specificity of the P2/NP/MP primer in comparison to 2550F/2718R primer, where both primers have not been demonstrated to successfully choose the gender of our buzzards. It was found that 2550F/2718R primer has 78% (7/9) specificity, whereas the P2/NP/MP primer showed 100% (9/9) specificity. Also, we found that male birds displayed a 600–700 base pairs fragment PCR products (depending on species), whereas females showed two fragments, one including 600–700 base pairs and another with 400–500 base pairs. To find the exact length of the fragments, cloning and nucleotide sequencing is needed. In conclusion, we demonstrate that the P2/NP/MP primer set was able identify the gender of a rufous-winged buzzard through PCR amplification, with high specificity and sensitivity. AcknowledgmentsThe authors would like to thank the Faculty of Agricultural Technology, Burapha University, Sakaeo Campus, for providing the equipment. They sincerely thank the students in the Faculty of Agricultural Technology at Burapha University, Sakaeo Campus, and animal husbandry officer from Watthana Nakhon district Livestock Office for the blood sample collection of rufous-winged buzzard. They are also grateful to John Morris of KMITL Research and Innovation Services (KRIS) for proofreading the article. Conflict of interestThe authors declare that there is no conflict of interest. Authors’ contributionsAll authors conceived the study, wrote the paper, and participated in monitoring the results. All authors read and approved the final manuscript. ReferencesChomczynsky, P. and Sacchi, N. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate phenol choroform extraction. Anal. Biochem. 162, 156–159. Costantini, V., Guaricci, A.C., Laricchiuta, P., Rausa, F. and Lacalandra, G.M. 2008. DNA sexing in Humboldt Penguins (Spheniscus humboldti) from feather samples. Anim. Reprod. Sci. 106, 162–167. Dubiec, A. and Magdalena, Z.N. 2006. Molecular techniques for sex identification in birds. Biol. Lett. 43, 3–12. Ellegren, H. 1996. First gene on the avian W chromosome (CHD) provides a tag for universal sexing of non-ratite birds. Proc. R. Soc. Lond. B Biol. Sci. 263, 1635–1641. Fridolfsson, A.K. and Ellegren, H. 1999. A simple and universal method for molecular sexing of non-ratite birds. J. Avian Biol. 20, 116–121. Garofalo, L., Fanelli, R., Opramolla, G., Polidori, M., Tancredi, F., Altea, T., Posillico, M. and Lorenzini, R. 2016. Comparison between two molecular protocols for sex determination in birds, with implications for the management and conservation of the Eurasian Griffon vulture Gyps fulvus. Avocetta 40, 17–22. Griffiths, R. and Tiwari, B. 1995. Sex of the last wild spix’s macaw. Nature 375, 454. Griffiths, R., Double, M.C., Orr, K. and Dawson, R.J.G. 1998. A DNA test to sex most birds. Mol. Ecol. 7, 1071–1075. Hammond, J.B.W., Spanswick, G. And Mawn, J.A. 1996. Extraction of DNA from preserved animal specimens for use in randomly amplified polymorphic DNA analysis. Anal. Biochem. 240, 298–300. Idaghdour, Y., Broderick, D. and Korrida, A. 2003. Faeces as a source of DNA for molecular studies in a threatened population of great bustards. Conserv. Genet. 4, 789–792. Ito, H., Yamaji, A.S., Abe, M., Murase, T. and Tsubota, T. 2003. Sex identification by alternative polymerase chain reaction methods in falconiformes. Zoo. Sci. 20, 339–344. Jensen, T., Pernasetti, F.M. and Durrant, B. 2003. Conditions for rapid sex determination in 47 avian species by PCR of genomic DNA from blood, shell-membrane blood vessels and feathers. Zoo. Biol. 22, 561–571. Kahn, N.W., John, J.S. and Quinn, T.W. 1998. Chromosome-specific intron size differences in the avian CHD gene provide an efficient method for sex identification in birds. Auk 115, 1074–1078. Lee, M.Y., Hong, Y.J., Park, S.K., Kim, Y.J., Choi, T.Y., Lee, H. and Min, M.S. 2008. Application of two complementary molecular sexing methods for east Asian bird species. Gene Genomics 30, 365–372. Prabhakar, B.G., Praveen, F.G., Vibhu, P., Richard, J.C., Mandar, K., Nikita, P., Asit, D., Anil, K.S. and Mohini, S. 2012. Molecular sexing of threatened Gyps vultures: an important strategy for conservation breeding and ecological studies. Springer Plus 1, 1–12. Reddy, A., Prakash, V. and Shivaji, S. 2007. A rapid, non-invasive, PCR-based method for identification of sex of the endangered old world vultures (white-backed and long-billed vultures)—implications for captive breeding programmes. Curr. Sci. 92, 659–662. Robertson, B.C., Minot, E.O. and Lambert, D.M. 1999. Molecular sexing of individual kakapo, Strigops habroptilus Aves, from faeces. Mol. Ecol. 8, 1349–1350. Sacchi, P., Soglia, D., Maione, S., Meneguz, G., Campora, M. and Rasero, R. 2004. A non-invasive test for sex identification in Short-toed Eagle (Circaetus gallicus). Mol. Cell Probes. 18, 193–196. Sambrook, J., Fritsch, E.F. and Maniatis, T. 1989. Molecular cloning: a laboratory manual, 2nd ed. New York, NY: Cold Spring Harbor Laboratory Press, p: 628. Thammakarn, C., Punchukrang, A., Jirajaroenrat, K. and Srikijkasemwat, K. 2007. Sex identification of some psittacine birds by polymerase chain reaction. J. Mahanakorn Vet. Med. 2, 30–34. Walsh, P.S., Metzger, D.A. and Higuchi, R. 1991. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques 10, 506–513. Wang, L.C., Chen, C.T., Lee, H.Y., Li, S.H., Lir, J.T., Chin, S.C., Pu, C.E. and Wang, C.H. 2007. Sexing a wider range of avian species based on two CHD1 introns with a unified reaction condition. Zoo. Biol. 26, 425–431. Watson, R.M., Griaznova, O.I., Long C.M. and. Holland, M.J. 2004. Increased sample capacity for genotyping and expression profiling by kinetic polymerase chain reaction. Anal. Biochem. 329, 58–67. | ||
| How to Cite this Article |
| Pubmed Style Kaewhom P, Srikijkasemwat K. Efficacy of two primer sets used in Sex Identification of Rufous-winged Buzzard (Butastur liventer).. Open Vet. J.. 2021; 11(4): 581-586. doi:10.5455/OVJ.2021.v11.i4.7 Web Style Kaewhom P, Srikijkasemwat K. Efficacy of two primer sets used in Sex Identification of Rufous-winged Buzzard (Butastur liventer).. https://www.openveterinaryjournal.com/?mno=91008 [Access: January 25, 2026]. doi:10.5455/OVJ.2021.v11.i4.7 AMA (American Medical Association) Style Kaewhom P, Srikijkasemwat K. Efficacy of two primer sets used in Sex Identification of Rufous-winged Buzzard
(Butastur liventer).. Open Vet. J.. 2021; 11(4): 581-586. doi:10.5455/OVJ.2021.v11.i4.7 Vancouver/ICMJE Style Kaewhom P, Srikijkasemwat K. Efficacy of two primer sets used in Sex Identification of Rufous-winged Buzzard
(Butastur liventer).. Open Vet. J.. (2021), [cited January 25, 2026]; 11(4): 581-586. doi:10.5455/OVJ.2021.v11.i4.7 Harvard Style Kaewhom, P. & Srikijkasemwat, . K. (2021) Efficacy of two primer sets used in Sex Identification of Rufous-winged Buzzard
(Butastur liventer).. Open Vet. J., 11 (4), 581-586. doi:10.5455/OVJ.2021.v11.i4.7 Turabian Style Kaewhom, Paitoon, and Kanokrat Srikijkasemwat. 2021. Efficacy of two primer sets used in Sex Identification of Rufous-winged Buzzard
(Butastur liventer).. Open Veterinary Journal, 11 (4), 581-586. doi:10.5455/OVJ.2021.v11.i4.7 Chicago Style Kaewhom, Paitoon, and Kanokrat Srikijkasemwat. "Efficacy of two primer sets used in Sex Identification of Rufous-winged Buzzard
(Butastur liventer).." Open Veterinary Journal 11 (2021), 581-586. doi:10.5455/OVJ.2021.v11.i4.7 MLA (The Modern Language Association) Style Kaewhom, Paitoon, and Kanokrat Srikijkasemwat. "Efficacy of two primer sets used in Sex Identification of Rufous-winged Buzzard
(Butastur liventer).." Open Veterinary Journal 11.4 (2021), 581-586. Print. doi:10.5455/OVJ.2021.v11.i4.7 APA (American Psychological Association) Style Kaewhom, P. & Srikijkasemwat, . K. (2021) Efficacy of two primer sets used in Sex Identification of Rufous-winged Buzzard
(Butastur liventer).. Open Veterinary Journal, 11 (4), 581-586. doi:10.5455/OVJ.2021.v11.i4.7 |